-Search query
-Search result
Showing 1 - 50 of 93 items for (author: weber & g)

EMDB-19562: 
Vimentin intermediate filament protofibril stoichiometry
Method: subtomogram averaging / : Eibauer M, Medalia O

EMDB-19563: 
Vimentin intermediate filament structure (delta tail)
Method: helical / : Eibauer M, Medalia O

EMDB-16844: 
Vimentin intermediate filament structure
Method: helical / : Eibauer M, Medalia O

EMDB-18953: 
Cryo-electron tomogram of Yersinia entomophaga chi2-sfGFP cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-18954: 
Cryo-electron tomogram of Yersinia entomophaga chi2-sfGFP cells
Method: electron tomography / : Feldmueller M, Pilhofer M
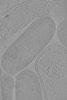
EMDB-18955: 
Cryo-electron tomogram of Yersinia entomophaga chi2-sfGFP cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-18957: 
Cryo-electron tomogram of Yersinia entomophaga MH96 cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-18958: 
Cryo-electron tomogram of Yersinia entomophaga MH96 cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-18960: 
Cryo-electron tomogram of Yersinia entomophaga delta LC cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-18961: 
Cryo-electron tomogram of mechanically cryo-milled Yersinia entomophaga delta LC cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-18962: 
Cryo-electron tomogram of a lysate preparation of Yersinia entomophaga delta LC cells
Method: electron tomography / : Feldmueller M, Pilhofer M
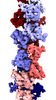
EMDB-18970: 
Subtomogram average of M66 filaments in Yersinia entomophaga cells
Method: subtomogram averaging / : Feldmueller M, Afanasyev P, Pilhofer M
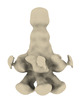
EMDB-18971: 
Subtomogram average of YenTc-Chi2-sfGFP from Yersinia entomophaga chi2-sfGFP
Method: subtomogram averaging / : Feldmueller M, Pilhofer M
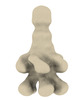
EMDB-18972: 
Subtomogram average of YenTc from Yersinia entomophaga MH96
Method: subtomogram averaging / : Feldmueller M, Pilhofer M

EMDB-19370: 
Cryo-electron tomogram of Yersinia entomophaga delta YenTc cells
Method: electron tomography / : Feldmueller M, Pilhofer M
EMDB-19371: 
Cryo-electron tomogram of Yersinia entomophaga delta YenTc cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-19372: 
Cryo-electron tomogram of Yersinia entomophaga delta YenTc cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-19373: 
Cryo-electron tomogram of Yersinia entomophaga delta YenTc cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-19374: 
Cryo-electron tomogram of Yersinia entomophaga chi2-sfGFP cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-19375: 
Cryo-electron tomogram of Yersinia entomophaga chi2-sfGFP cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-19376: 
Cryo-electron tomogram of Yersinia entomophaga delta LC delta YenTc cells
Method: electron tomography / : Feldmueller M, Pilhofer M
EMDB-19377: 
Cryo-electron tomogram of Yersinia entomophaga MH96 cells grown at 37 degrees
Method: electron tomography / : Feldmueller M, Pilhofer M
EMDB-19378: 
Cryo-electron tomogram of Yersinia entomophaga delta LC cells grown at 37 degrees
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-19379: 
Cryo-electron tomogram of Yersinia entomophaga delta LC delta M66 cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-19380: 
Cryo-electron tomogram of Yersinia entomophaga delta LC delta M66 cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-19381: 
Cryo-electron tomogram of a lysate preparation of Yersinia entomophaga delta LC delta M66 cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-16024: 
SARS-CoV-2 S protein in complex with pT1644 Fab
Method: single particle / : Stroeh L, Hansen G, Vollmer B, Krey T, Benecke T, Gruenewald K

EMDB-16026: 
SARS-CoV-2 S protein in complex with pT1696 Fab
Method: single particle / : Hansen G, Benecke T, Vollmer B, Gruenewald K, Krey T

EMDB-28205: 
Clostridioides difficile binary toxin translocase CDTb wild-type after calcium depletion from receptor binding domain 1 (RBD1) - Class 2
Method: single particle / : Abeyawardhane DL, Pozharski E

EMDB-28206: 
Clostridioides difficile binary toxin translocase CDTb wild-type after calcium depletion from receptor binding domain 1 (RBD1) - Class 1
Method: single particle / : Abeyawardhane DL, Pozharski E

EMDB-28207: 
Clostridioides difficile binary toxin translocase CDTb double mutant - D623A D734A
Method: single particle / : Abeyawardhane DL, Pozharski E
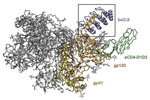
EMDB-26157: 
Cryo-EM structure of BG505 SOSIP HIV-1 Env trimer in complex with CD4 receptor (D1D2) and broadly neutralizing darpin bnD.9
Method: single particle / : Cerutti G, Gorman J

PDB-7txd: 
Cryo-EM structure of BG505 SOSIP HIV-1 Env trimer in complex with CD4 receptor (D1D2) and broadly neutralizing darpin bnD.9
Method: single particle / : Cerutti G, Gorman J, Kwong PD, Shapiro L

EMDB-15987: 
In situ sub-tomogram average of the Ca. L. ossiferum ribosome
Method: subtomogram averaging / : Rodrigues-Oliveira T, Wollweber F, Ponce-Toledo R, Xu J, Rittmann S, Klingl A, Pilhofer M, Schleper C

EMDB-15988: 
In situ sub-tomogram average of Ca. L. ossiferum actin filament
Method: subtomogram averaging / : Rodrigues-Oliveira T, Wollweber F, Ponce-Toledo R, Xu J, Rittmann S, Klingl A, Pilhofer M, Schleper C

EMDB-15989: 
cryo-tomogram of a Ca. L. ossiferum cell
Method: electron tomography / : Rodrigues-Oliveira T, Wollweber F, Ponce-Toledo R, Xu J, Rittmann S, Klingl A, Pilhofer M, Schleper C

EMDB-15990: 
cryo-tomogram of Ca. L. ossiferum protrusions
Method: electron tomography / : Rodrigues-Oliveira T, Wollweber F, Ponce-Toledo R, Xu J, Rittmann S, Klingl A, Pilhofer M, Schleper C

EMDB-15991: 
cryo-tomogram of a Ca.L. ossiferum cell
Method: electron tomography / : Rodrigues-Oliveira T, Wollweber F, Ponce-Toledo R, Xu J, Rittmann S, Klingl A, Pilhofer M, Schleper C

EMDB-15992: 
cryo-tomogram of a Ca. L. ossiferum cell
Method: electron tomography / : Rodrigues-Oliveira T, Wollweber F, Ponce-Toledo R, Xu J, Rittmann S, Klingl A, Pilhofer M, Schleper C

EMDB-15993: 
cryo-tomogram of a Ca. L. ossiferum cell
Method: electron tomography / : Rodrigues-Oliveira T, Wollweber F, Ponce-Toledo R, Xu J, Rittmann S, Klingl A, Pilhofer M, Schleper C

EMDB-12589: 
Plasmodium falciparum Glutamine synthetase (PfGs)
Method: single particle / : Makumire S, Mlelu AE, Gqunu S, Weber WB, Sewell BT

EMDB-13045: 
Cryo-EM structure of Brr2 in complex with Fbp21
Method: single particle / : Bergfort A, Hilal T, Weber G, Wahl MC

EMDB-13046: 
Cryo-EM structure of Brr2 in complex with Jab1/MPN and C9ORF78
Method: single particle / : Bergfort A, Hilal T, Weber G, Wahl MC

PDB-7os1: 
Cryo-EM structure of Brr2 in complex with Fbp21
Method: single particle / : Bergfort A, Hilal T, Weber G, Wahl MC

PDB-7os2: 
Cryo-EM structure of Brr2 in complex with Jab1/MPN and C9ORF78
Method: single particle / : Bergfort A, Hilal T, Weber G, Wahl MC

EMDB-13690: 
Structure of U5 snRNP assembly and recycling factor TSSC4 in complex with BRR2 and Jab1 domain of PRPF8
Method: single particle / : Bergfort A, Kuropka B, Ilik IA, Freund C, Aktas T, Hilal T, Weber G, Wahl MC

PDB-7px3: 
Structure of U5 snRNP assembly and recycling factor TSSC4 in complex with BRR2 and Jab1 domain of PRPF8
Method: single particle / : Bergfort A, Kuropka B, Ilik IA, Freund C, Aktas T, Hilal T, Weber G, Wahl MC

EMDB-13204: 
transcriptional activator PafBC bound to mycobacterial RNA polymerase
Method: single particle / : Mueller AU, Kummer E, Schilling CM, Ban N, Weber-Ban E

EMDB-13205: 
Mycobacterial RNAP with transcriptional activator PafBC
Method: single particle / : Mueller AU, Kummer E, Schilling CM, Ban N, Weber-Ban E

PDB-7p5x: 
Mycobacterial RNAP with transcriptional activator PafBC
Method: single particle / : Mueller AU, Kummer E, Schilling CM, Ban N, Weber-Ban E
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
